
A new method, described in a study published yesterday in the journal Nature Communications, has the potential to boost international research efforts to find drugs that eradicate cancer at its source.
Most cancerous tissue consists of rapidly dividing cells with a limited capacity for self-renewal, meaning that the bulk of cells stop reproducing after a certain number of divisions. However, cancer stem cells can replicate indefinitely, fuelling long-term cancer growth and driving relapse.
Cancer stem cells that elude conventional treatments like chemotherapy are one of the reasons patients initially enter remission but relapse soon after. In acute myeloid leukaemia, a form of blood cancer, the high probability of relapse means fewer than 15% of elderly patients live longer than five years.
However, cancer stem cells are difficult to isolate and study because of their low abundance and similarity to other stem cells, hampering international research efforts in developing precision treatments that target malignant cells while sparing healthy ones.
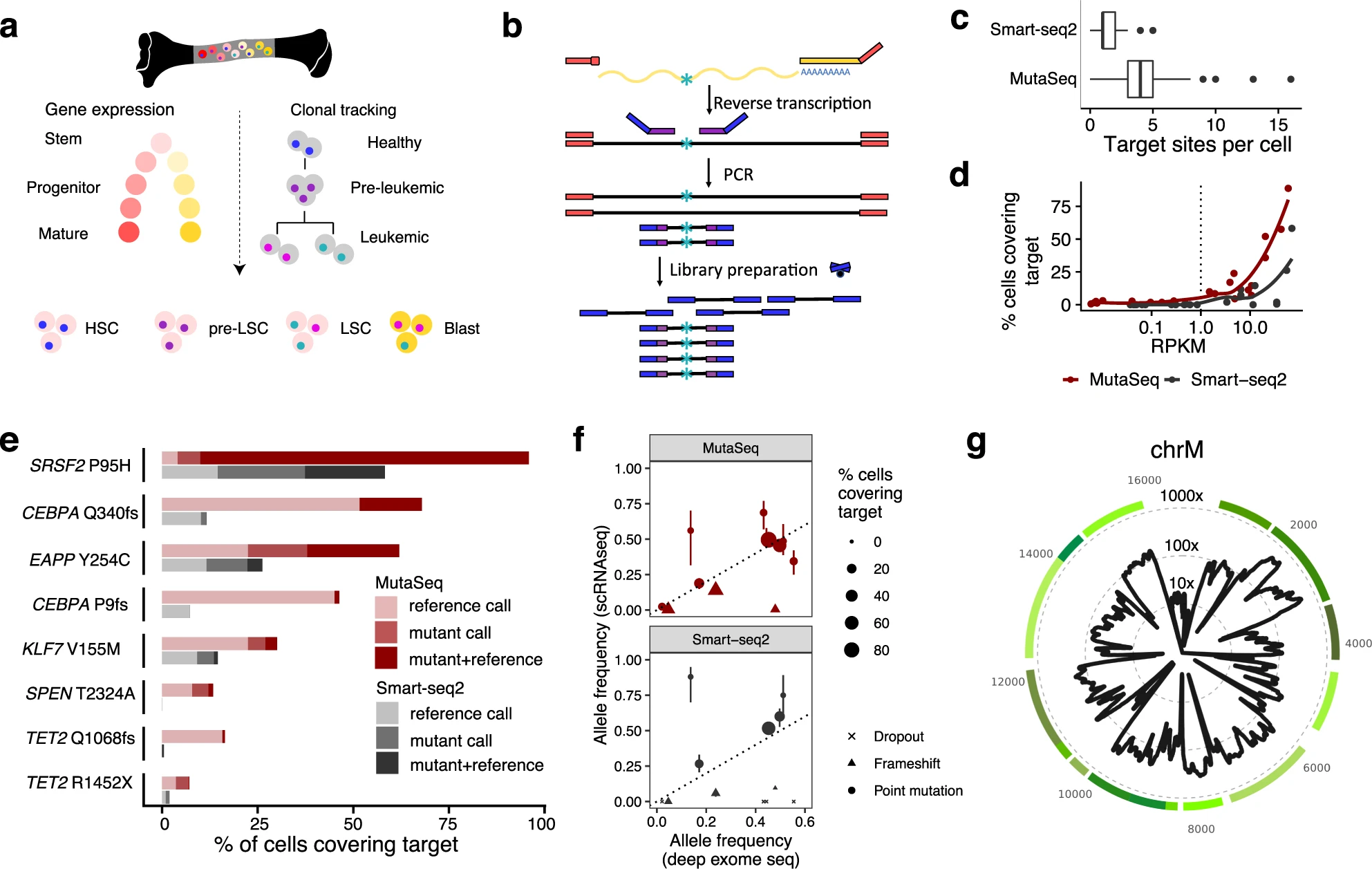
Researchers from the Centre for Genomic Regulation (CRG) and the European Molecular Biology Laboratory (EMBL) have overcome this problem by creating MutaSeq, a method that can be used to distinguish cancer stem cells, mature cancer cells and otherwise healthy stem cells based on their genetics and gene expression.
“RNA provides vital information for human health. For example, PCR tests for coronavirus detect its RNA to diagnose COVID-19. Subsequent sequencing can determine the virus variant,” explains Lars Velten, Group Leader at the CRG and author of the paper. “MutaSeq works like a PCR test for coronavirus, but at a much more complex level and with a single cell as starting material.”


To determine if a single cell is a stem cell, the researchers used MutaSeq to measure thousands of RNAs at the same time. To then find out if the cell is cancerous or healthy, the researchers carried out additional sequencing and looked for mutations. The resulting data helped researchers track if stems cells are cancerous or healthy and helped determine what makes the cancer stem cells different.
“There are a huge number of small molecule drugs out there with demonstrated clinical safety, but deciding which cancers and more specifically which patients these drugs are well suited for is a daunting task,” says Lars Steinmetz, Professor at Stanford University, Group Leader at EMBL Heidelberg and author of the paper. “Our method can identify drug targets that might not have been tested in the right context. These tests will need to be carried out in controlled clinical studies, but knowing what to try is an important first step.”
The method is based on single cell sequencing, an increasingly common technique that helps researchers gather and interpret genome-wide information from thousands of individual cells. Single cell sequencing provides a highly detailed molecular profile of complex tissues and cancers, opening new avenues for research.
Explaining their next steps, Lars Velten says: “We have now brought together clinical researchers from Germany and Spain to apply this method in much larger clinical studies. We are also making the method much more streamlined. Our vision is to identify cancer stem cell specific drug targets in a personalized manner, making it ultimately as easy for patients and doctors to look for these treatments as it is testing for coronavirus”.

