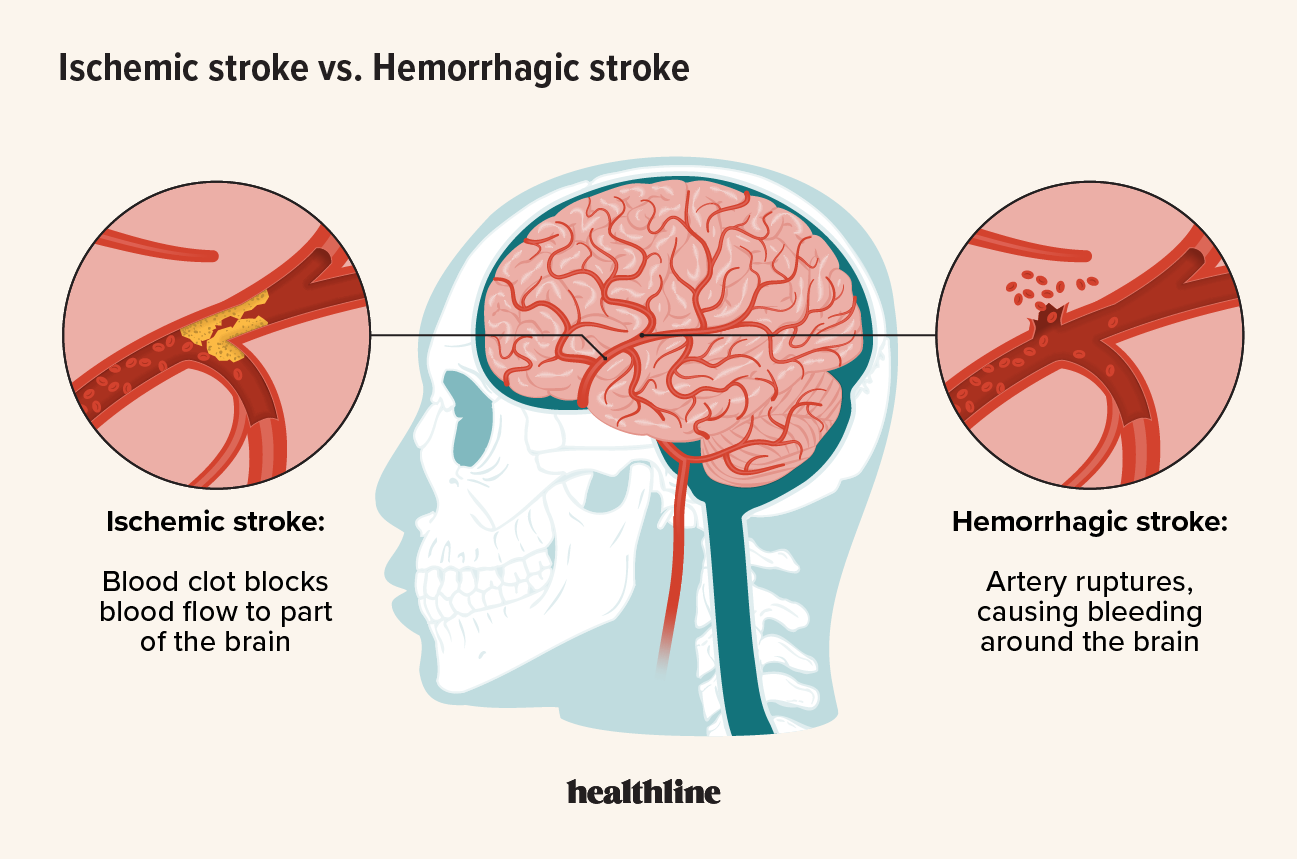

The study discovered genetic markers in inflammation that may be related to a second stroke or other major cardiovascular event following a stroke. These findings could help identify drug targets to mitigate stroke-related disability and mortality.
People who experience an arterial ischemic stroke (AIS) or transient ischemic stroke (TIA) are at an increased risk of suffering a second stroke or other major adverse cardiovascular event (MACE), making it critically important to identify risk factors and treatments to prevent these subsequent occurrences.
A new study led by Boston University School of Public Health (BUSPH), the National Institute for Health and Care Research (NIHR) Bristol Biomedical Research Centre (Bristol BRC), and Veteran’s Affairs Boston Healthcare System (VA Boston), has identified new genetic and molecular risk factors that may reveal new pathways for treating patients after they experience their first stroke.
Published in Stroke, a journal of the American Heart Association, the study identified CCL27 and TNFRSF14, two proteins that are associated with subsequent MACE, but not initial strokes. These proteins are known to activate inflammation, which plays a key role in the development of strokes and many chronic conditions and diseases. The findings suggest that inflammation is a contributing factor to MACE outcomes among people after they have their first stroke.
“While previous studies have found associations between inflammation and incident AIS/MACE, our study found that these causal proteins may also have a role in subsequent MACE, which could lead to potential novel drug targets,” says study co-lead author Nimish Adhikari, a PhD student in biostatistics at BUSPH and VA Boston. The study was also co-led by Andrew Elmore, senior research associate in health data science at NIHR Bristol BRC.
Utilizing genetic information and medical history data from two large biobanks, the VA’s Million Veteran Program and UK Biobank, the research team conducted ancestry-specific genome-wide association studies (GWAS) to find associations between DNA and incident and subsequent AIS and MACE.
GWAS are typically performed to determine whether individuals have had a medical event for the first time, but applying this method to subsequent MACE events could shed novel insights about stroke progression, information that would be valuable for therapeutic drug identification, the researchers say.
In total, the researchers examined 93,422 individuals who had an incident stroke, among which 51,929 had subsequent MACE and 45,120 had subsequent AIS.
In population specific analyses, they observed two significant genetic variants: rs76472767, near gene RNF220 on chromosome 1 in the African ancestry GWAS for subsequent MACE, and rs13294166, near gene LINC01492 on chromosome 9 in the same ancestry GWAS for subsequent AIS.
“We used that data to find if there were certain molecules that were associated with either incident or subsequent states,” says Elmore. “From that, we were able to identify a link between certain molecules that play a part in inflammation and these stroke and MACE outcomes.”
While the prevalence of stroke has declined worldwide over the last three decades, it is still the second-leading cause of death and third-leading cause of disability across the globe, and it remains a significant public health issue. Stroke also continues to disproportionately affect populations among racial, ethnic, socioeconomic, and geographical lines, furthering health inequities in both high- and low-income countries. Identifying novel drug targets for new therapeutic interventions that thwart stroke progression could save millions of people from experiencing stroke-related disability and mortality.
It’s unknown if targeting other modifiable risk factors for stroke could also offer pathways for effective treatment after someone experiences their first stroke.
“We are looking forward to extending this research to other cardiometabolic outcomes beyond stroke,” says co-senior and corresponding author Gina Peloso, associate professor of biostatistics at BUSPH.

